Gaussian Software Free Download Updated Version 2025
Gaussian 16 is the latest version of the Gaussian series of electronic structure programs, used by chemists, chemical engineers, biochemists, physicists, and other scientists worldwide. Gaussian 16 provides a wide-ranging suite of the most advanced modeling capabilities available. You can use it to investigate the real-world chemical problems that interest you, in all of their complexity, even on modest computer hardware. Starting from the fundamental laws of quantum mechanics, Gaussian software 16 predicts the energies, molecular structures, vibrational frequencies, and molecular properties of compounds and reactions in a wide variety of chemical environments. Gaussian 16’s models can be applied to both stable species and compounds that are difficult or impossible to observe experimentally, whether due to their nature (e.g., toxicity, combustibility, radioactivity) or their inherent fleeting nature (e.g., short-lived intermediates and transition structures). Gaussian 16 can predict a variety of spectra, in both the gas phase and in solution, including IR and Raman, NMR spectra and spin-spin coupling constants, vibrational circular dichroism (VCD), Raman optical activity (ROA), resonance Raman, UV/Visible, vibronic absorption and emission spectra for excited states via Franck-Condon and/or Herzberg-Teller analysis, electronic circular dichroism (ECD) and circularly polarized luminescence (CPL), optical rotatory dispersion (ORD), and hyperfine (microwave spectroscopy). Anharmonic analysis is available for IR, Raman, VCD, and ROA spectra.

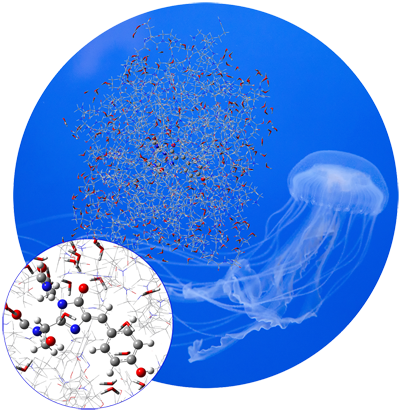
Continuing the nearly 40-year tradition of the Gaussian series of electronic structure programs, Gaussian 16 offers new methods and capabilities that allow you to study ever larger molecular systems and additional areas of chemistry. GaussView 6 offers a rich set of building and visualization capabilities. We highlight some of the most important features on this page. GFP is a protein that fluoresces bright green when exposed to light in the blue-to-ultraviolet range. The chromophore is shown in the inset below. The molecule was first isolated in the jellyfish species Aequorea victoria, which is native to the Pacific Northwest coast of North America. Since then, it has been studied extensively, and variants of the molecule with enhanced fluorescence properties have been engineered.GFP consists of a chromophore within a protein chain composed of 238 amino acids. The isolated chromophore is not fluorescent, so modeling it in its protein environment is essential. GFP’s fluorescence cycle involves an initial excitation to its first excited state, a proton transfer reaction on the S1 potential energy surface, and finally a relaxation back to the ground state.
The following features of Gaussian software 16 and GaussView 6 are useful for modeling fluorescence in this compound.
GaussView can directly open files from the Protein Data Bank (PDB files). It can add hydrogens to the retrieved structure when imported or at a later time. You can also view, manipulate, and modify the structure using the PDB chain, residue type and/or number, and other PDB substructure information present in the original file. Residue and other information can be retained throughout molecule editing and job execution.
The molecule can be modeled via MO:MM calculations using Gaussian’s ONIOM facility:
- GaussView makes it easy to define ONIOM layers based on many different criteria. For this molecule, assigning atoms by PDB residue is often the most straightforward.
- GaussView identifies molecular mechanics atom types and partial charges automatically in most cases. It is also simple to locate atoms missing types and to specify/modify types and charges as desired. Gaussian incorporates standard MM parameters for Amber and other force fields and also allows you to define MM parameters as needed.
- Gaussian’s ONIOM electronic embedding feature includes all of the effects of the protein environment without neglecting terms in the MM coupling with the chromophore QM treatment.
- Explicit solvent molecules can be included in the calculation (e.g., water molecules above).
- GaussView can directly open files from the Protein Data Bank (PDB files). It can add hydrogens to the retrieved structure when imported or at a later time. You can also view, manipulate, and modify the structure using the PDB chain, residue type and/or number, and other PDB substructure information present in the original file. Residue and other information can be retained throughout molecule editing and job execution.
- The molecule can be modeled via MO:MM calculations using Gaussian’s ONIOM facility:
- GaussView makes it easy to define ONIOM layers based on many different criteria. For this molecule, assigning atoms by PDB residue is often the most straightforward.
- GaussView identifies molecular mechanics atoms types and partial charges automatically in most cases. It is also simple to locate atoms missing types and to specify/modify types and charges as desired. Gaussian incorporates standard MM parameters for Amber and other force fields and also allows you to define MM parameters as needed.
- Gaussian’s ONIOM electronic embedding feature includes all of the effects of the protein environment without neglecting terms in the MM coupling with the chromophore QM treatment.
- Explicit solvent molecules can be included in the calculation (e.g., water molecules above).
- GaussView can directly open files from the Protein Data Bank (PDB files). It can add hydrogens to the retrieved structure when imported or at a later time. You can also view, manipulate, and modify the structure using the PDB chain, residue type and/or number, and other PDB substructure information present in the original file. Residue and other information can be retained throughout molecule editing and job execution.
:
Gaussian software 16 Features at a Glance
Features introduced since Gaussian software 09 Rev A are in blue.
Existing features enhanced in Gaussian 16 are in green.
Fundamental Algorithms
- Calculation of one- & two-electron integrals over any contracted gaussian functions
- Conventional, direct, semi-direct and in-core algorithms
- Linearized computational cost via automated fast multipole methods (FMM) and sparse matrix techniques
- Harris initial guess
- Initial guess generated from fragment guesses or fragment SCF solutions
- Density fitting and Coulomb engine for pure DFT calculations, including automated generation of fitting basis sets
exact exchange for HF and hybrid DFT
- 1D, 2D, 3D periodic boundary conditions (PBC) energies & gradients (HF & DFT)
- Shared-memory (SMP), cluster/network and GPU-based parallel execution
Model Chemistries
Molecular Mechanics
- Amber, DREIDING and UFF energies, gradients, and frequencies
- Custom force fields
- Standalone MM program
Ground State Semi-Empirical
- CNDO/2, INDO, MINDO3 and MNDO energies and gradients
- AM1, PM3, PM3MM, PM6 and PDDG energies, gradients and reimplemented (analytic) frequencies
- PM7: original and modified for continuous potential energy surfaces
- Custom semi-empirical parameters (Gaussian and MOPAC External formats)
- DFTB and DFTBA methods
Self Consistent Field (SCF)
- SCF restricted and unrestricted energies, gradients and frequencies, and RO energies and gradients
- EDIIS+CDIIS default algorithm; optional Quadratic Convergent SCF
- SCF procedure enhancements for very large calculations
- Complete Active Space SCF (CASSCF) energies, gradients & frequencies
- Active spaces of up to 16 orbitals
- Restricted Active Space SCF (RASSCF) energies and gradients
- Generalized Valence Bond-Perfect Pairing energies and gradients
- Wavefunction stability analysis (HF & DFT)
Density Functional Theory
Closed and open shell energies, gradients & frequencies, and RO energies & gradients are available for all DFT methods.
- EXCHANGE FUNCTIONALS: Slater, Xα, Becke 88, Perdew-Wang 91, Barone-modified PW91, Gill 96, PBE, OPTX, TPSS, revised TPSS, BRx, PKZB, ωPBEh/HSE, PBEh
- CORRELATION FUNCTIONALS: VWN, VWN5, LYP, Perdew 81, Perdew 86, Perdew-Wang 91, PBE, B95, TPSS, revised TPSS, KCIS, BRC, PKZB, VP86, V5LYP
- OTHER PURE FUNCTIONALS: VSXC, HCTH functional family, τHCTH, B97D, M06L, SOGGA11, M11L, MN12L, N12, MN15L
- HYBRID METHODS: B3LYP, B3P86, P3PW91, B1 and variations, B98, B97-1, B97-2, PBE1PBE, HSEh1PBE and variations, O3LYP, TPSSh, τHCTHhyb, BMK, AFD, M05, M052X, M06, M06HF, M062X, M08HX, PW6B95, PW6B95D3, M11, SOGGA11X, N12, MN12SX, N12SX, MN15, HISSbPBE, X3LYP, BHandHLYP; user-configurable hybrid methods
- DOUBLE HYBRID: B2PLYP & mPW2PLYP and variations with dispersion, DSDPBEP86, PBE0DH, PBEQIDH (see also below in “Electron Correlation”)
- EMPIRICAL DISPERSION: PFD, GD2, GD3, GD3BJ
- FUNCTIONALS INCLUDING DISPERSION: APFD, B97D3, B2PLYPD3
- LONG RANGE-CORRECTED: LC-ωPBE, CAM-B3LYP, ωB97XD and variations, Hirao’s general LC correction
- Larger numerical integrations grids
Electron Correlation:
All methods/job types are available for both closed and open shell systems and may use frozen core orbitals; restricted open shell calculations are available for MP2, MP3, MP4 and CCSD/CCSD(T) energies.
- MP2 energies, gradients, and frequencies
- Double hybrid DFT energies, gradients and frequencies, with optional empirical dispersion (see list in “Density Functional Theory” above)
- CASSCF calculations with MP2 correlation for any specified set of states
- MP3 and MP4(SDQ) energies and gradients
- MP4(SDTQ) and MP5 energies
- Configuration Interaction (CISD) energies & gradients
- Quadratic CI energies & gradients; QCISD(TQ) energies
- Coupled Cluster methods: restartable CCD, CCSD energies & gradients, CCSD(T) energies; optionally input amplitudes computed with smaller basis set
- Optimized memory algorithm to avoid I/O during CCSD iterations
- Brueckner Doubles (BD) energies and gradients, BD(T) energies; optionally input amplitudes & orbitals computed with a smaller basis set
- Enhanced Outer Valence Green’s Function (OVGF) methods for ionization potentials & electron affinities
- Complete Basis Set (CBS) MP2 Extrapolation
- Douglas-Kroll-Hess scalar relativistic Hamiltonians
Automated High Accuracy Energies
- G1, G2, G3, G4 and variations
- CBS-4, CBS-q, CBS-QB3, ROCBS-QB3, CBS-Q, CBS-APNO
- W1U, W1BD, W1RO (enhanced core correlation energy calculation)
Basis Sets and DFT Fitting Sets
- STO-3G, 3-21G, 6-21G, 4-31G, 6-31G, 6-31G†, 6-311G, D95, D95V, SHC, CEP-nG, LanL2DZ, cc-pV{D,T,Q,5,6}Z, Dcc-p{D,T}Z, SV, SVP, TZV, QZVP, EPR-II, EPR-III, Midi!, UGBS*, MTSmall, DG{D, T}ZVP, CBSB7
- Augmented cc-pV*Z schemes: Aug- prefix, spAug-, dAug-, Truhlar calendar basis sets (original and regularized)
- Effective Core Potentials (through second derivatives): LanL2DZ, CEP through Rn, Stuttgart/Dresden
- Support for basis functions and ECPs of arbitrary angular momentum
- DFT FITTING SETS: DGA1, DGA1, W06, older sets designed for SVP and TZVP basis sets; auto-generated fitting sets; optional default enabling of density fitting
Geometry Optimizations and Reaction Modeling
- Geometry optimizations for equilibrium structures, transition structures, and higher saddle points, in redundant internal, internal (Z-matrix), Cartesian, or mixed internal and Cartesian coordinates
- GEDIIS optimization algorithm
- Redundant internal coordinate algorithm designed for large system, semi-empirical optimizations
- Newton-Raphson and Synchronous Transit-Guided Quasi-Newton (QST2/3) methods for locating transition structures
- IRCMax transition structure searches
- Relaxed and unrelaxed potential energy surface scans
- Implementation of intrinsic reaction path following (IRC), applicable to ONIOM QM:MM with thousands of atoms
- Reaction path optimization
- BOMD molecular dynamics (all analytic gradient methods); ADMP molecular dynamics: HF, DFT, ONIOM(MO:MM)
- Optimization of conical intersections via state-averaged CASSCF
- Generalized internal coordinates for complex optimization constraints
Vibrational Frequency Analysis
- Vibrational frequencies and normal modes (harmonic and anharmonic), including display/output limiting to specified atoms/residues/modes (optional mode sorting)
- Restartable analytic HF and DFT frequencies
- MO:MM ONIOM frequencies including electronic embedding
- Analytic Infrared and static and dynamic Raman intensities (HF & DFT; MP2 for IR)
- Pre-resonance Raman spectra (HF and DFT)
- Projected frequencies perpendicular to a reaction path
- NMR shielding tensors & GIAO magnetic susceptibilities (HF, DFT, MP2) and enhanced spin-spin coupling (HF, DFT)
- Vibrational circular dichroism (VCD) rotational strengths (HF and DFT; harmonic and anharmonic)
- Dynamic Raman Optical Activity (ROA) intensities (harmonic and anharmonic)
- Raman and ROA intensities calculated separately from force constants in order to use a larger basis set
- Harmonic vibration-rotation coupling
- Enhanced anharmonic vibrational analysis, including IR intensities, DCPT2 & HDCPT2 method for resonance-free computations of anharmonic frequencies
- Anharmonic vibration-rotation coupling via perturbation theory
- Hindered rotor analysis
Molecular Properties
- Population analysis, including per-orbital analysis for specifed orbitals: Mulliken, Hirshfeld, CM5
- Computed atomic charges can be saved for use in a later MM calculation
- Electrostatic potential, electron density, density gradient, Laplacian, and magnetic shielding & induced current densities over an automatically generated grid
- Multipole moments through hexadecapole
- Biorthogonalization of MOs (producing corresponding orbitals)
- Electrostatic potential-derived charges (Merz-Singh-Kollman, CHelp, CHelpG, Hu-Lu-Yang)
- Natural orbital analysis and natural transition orbitals
- Natural Bond Orbital (NBO) analysis, including orbitals for CAS jobs. Integrated support for NBO3; external interface to NBO6
- Static and frequency-dependent analytic polarizabilities and hyperpolarizabilities (HF and DFT); numeric 2nd hyperpolarizabilities (HF; DFT w/ analytic 3rd derivs.)
- Approx. CAS spin orbit coupling between states
- Enhanced optical rotations and optical rotary dispersion (ORD)
- Hyperfine spectra components: electronic g tensors, Fermi contact terms, anisotropic Fermi contact terms, rotational constants, dipole hyperfine terms, quartic centrifugal distortion, electronic spin rotation tensors, nuclear electric quadrupole constants, nuclear spin rotation tensors
- ONIOM integration of electric and magnetic properties
ONIOM Calculations
- Enhanced 2 and 3 layer ONIOM energies, gradients and frequencies using any available method for any layer
- Optional electronic embedding for MO:MM energies, gradients and frequencies implemented so as to include all effects of the MM environment without neglecting terms in its coupling with the QM region
- Enhanced MO:MM ONIOM optimizations to minima and transition structures via microiterations including electronic embedding
- Support for IRC calculations
- ONIOM integration of electric and magnetic properties
Excited States
- ZINDO energies
- CI-Singles energies, gradients, & freqs.
- Restartable time-dependent (TD) HF & DFT energies, gradients and frequencies. TD-DFT can use the Tamm-Dancoff approximation.
- SAC-CI energies and gradients
- EOM-CCSD energies and gradients (restartable); optionally input amplitudes computed with a smaller basis set
- Franck-Condon, Herzberg-Teller and FCHT analyses
- Vibronic spectra including electronic circular dichroism (ECD) rotational strengths (HF and DFT)
- Resonance Raman spectra
- Ciofini’s excited state charge transfer diagnostic (Dct)
- Caricato’s EOMCC solvation interaction models
- CI-Singles and TD-DFT in solution
- State-specific excitations and de-excitations in solution
- An energy range for excitations can be specified for CIS and TD excitation energies
Self-Consistent Reaction Field Solvation Models
- New implementation of the Polarized Continuum Model (PCM) facility for energies, gradients and frequencies
- Solvent effects on vibrational spectra, NMR, and other properties
- Solvent effects for ADMP trajectory calcs.
- Solvent effects for ONIOM calculations
- Enhanced solvent effects for excited states
- SMD model for ΔG of solvation
- Other SCRF solvent models (HF & DFT): Onsager energies, gradients and freqs., Isodensity Surface PCM (I-PCM) energies and Self-Consistent Isodensity Surface PCM (SCI-PCM) energies and gradients
Ease-of-Use Features
- Automated counterpoise calculations
- Automated optimization followed by frequency or single point energy
- Ability to easily add, remove, freeze, differentiate redundant internal coords.
- Simplified isotope substitution and temperature/pressure specification in the route section
- Optimizations
- Retrieve the nth geometry from a checkpoint file
- Recompute the force constants every nth step of a geometry optimization
- Reduce the maximum number of allowed steps, including across restarts
- 180° flips detected and suppressed for better visualization
- Freezing by fragment for ONIOM optimizations
- Simplified fragment definitions on molecule specifications
- Many more restartable job types
- Atom freezing in optimizations by type, fragment, ONIOM layer and/or residue
- QST2/QST3 automated transition structure optimizations
- Saving and reading normal modes
- %OldChk Link 0 command specifies read-only checkpoint file for data retrieval
- Default.Route file for setting calculation defaults
- Enhanced set of equivalent Default.Route directives, Link 0 commands, command line options and environment variables
Integration with External Programs
- NBO 6
- COSMO/RS
- AIMPAC WfnX files
- Antechamber
- ACID
- Pickett’s program
- DFTB input file
- General external interface script-based automation, results post-processing, interchanging data/calculation results with other programs, and so on:
- Interface routines in Fortran, Python and Perl (open source)
Computer Requirements: UNIX, Linux, macOS
Important Notes for All Gaussian Binary Versions:
• Beginning with Revision C.01, Linda 9.2 is required for network parallel use of Gaussian 16. Earlier versions of Linda are
incompatible. Thus, upgrading from Gaussian 16 Rev. A.03 or B.01 w/Linda to G16 Rev. C.01 or C.02 w/Linda or G16 Rev.
C.02 w/Linda requires an update to Linda 9.2.
• The Linux RedHat and SuSE versions specified for x86_64, IA32, and IBM Power systems refer only to the unmodified,
unpatched original media/ISO image distributions as released by the vendor.
Revision C.01 x86_64 AVX-enabled Binary Version:
• The AMD/Intel x86_64 AVX-enabled binary version includes GPU support for NVIDIA K40, K80, P100, and V100 boards
with 12 GB of memory or higher. A version of NVIDIA drivers compatible with CUDA 10.0 is required to run on these GPUs
with this binary version. (NVIDIA drivers can be found at www.nvidia.com/download/index.aspx).
.
Downlaod The File Gaussian Software
Download Gaussian Software for advanced molecular modeling and quantum chemistry calculations. It enables accurate simulations of molecular properties, reactions, and spectroscopy for cutting-edge research.